REPRESENTATIVE PAPERS

Sex-specific role of the 5-HT2A receptor in psilocybin-induced extinction of opioid reward
Nature Communications 2025

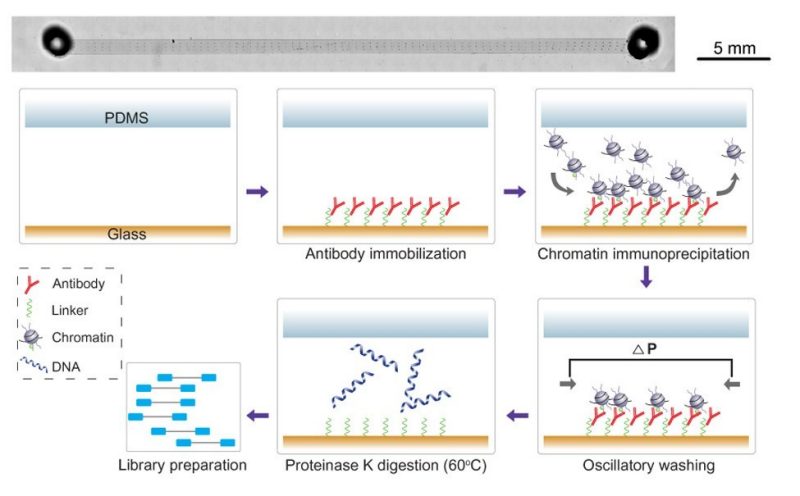
Epigenomic tomography for probing spatially defined chromatin state in the brain
Cell Reports Methods 2024
Liu, Z.#, Deng, C.#, Zhou, Z., Xiao, Y., Jiang, S., Zhu, B., Naler, L., Jia, X., Yao, D., Lu, C. Cell Reports Methods 4 (2024) 100738 Link

Antipsychotic-induced epigenomic reorganization in frontal cortex of individuals with schizophrenia
eLife 2023
Zhu, B.#, Ainsworth, R.J.#, Wang, Z., Liu, Z., Sierra, S., Deng, C., Callado, L.F., Meana, J.J., Wang, W.*, Lu, C.*, Gonzalez-Maeso, J*. eLife 12 (2023) RP92393. Link
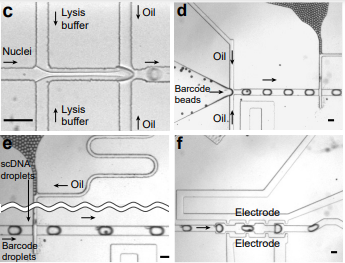
Droplet-based bisulfite sequencing for high throughput profiling of single-cell DNA methylomes
Nature Communications 2023
Zhang, Q., Ma, S., Liu, Z., Zhu, B., Zhou, Z., Li, G., Meana, J., González-Maeso, J., Lu, C. Nature Communications 14 (2023) 4672. Link
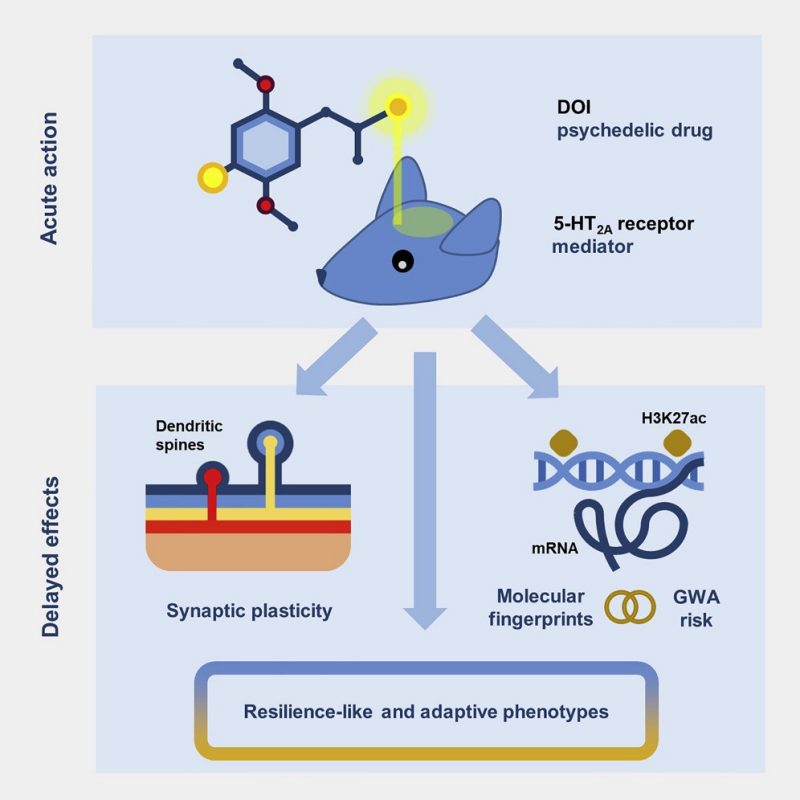
Prolonged epigenomic and synaptic plasticity alterations following single exposure to a psychedelic in mice
Cell Reports 2021
de la Fuente Revenga, M., Zhu, B., Guevara, C.A., Naler, L.B., Saunders, J.M., Zhou, Z., Toneatti, R., Sierra, S., Wolstenholme, J.T., Beardsley, P.M., Huntley, G.W., Lu, C.*, Gonzalez-Maeso, J.* Cell Reports 37 (2021) 109836. Link
MOWChIP-seq for low-input and multiplexed profiling of genome-wide histone modifications
Nature Protocols 2019
Zhu, B. , Hsieh, Y.-P. , Murphy, T. W., Zhang, Q., Naler, L. B., Lu, C. Nature Protocols 14 (2019) 3366-3394. Link
Low-input and multiplexed microfluidic assay reveals epigenomic variation across cerebellum and prefrontal cortex
Science Advances 2018
Ma, S., Hsieh, Y.-P., Ma, J., Lu, C. Science Advances 4 (2018) eaar8187. Link
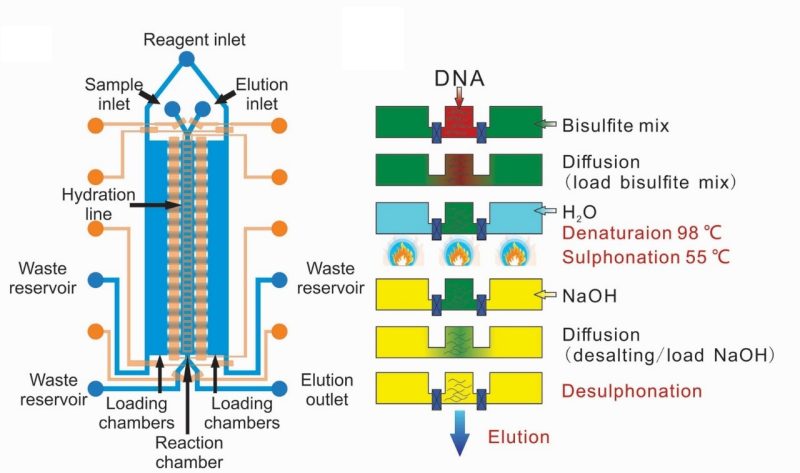
Cell-type-specific brain methylomes profiled via ultralow-input microfluidics
Nature Biomedical Engineering 2018
Ma, S., de la Fuente Revenga, M., Sun, Z., Sun, C., Murphy, T.W., Xie, H., Gonzalez-Maeso, J., Lu, C. Nature Biomedical Engineering 2 (2018) 183-194. Link; free view-only copy.
A microfluidic device for epigenomic profiling using 100 cells
Nature Methods 2015
Cao, Z., Chen, C., He, B., Tan, K., Lu, C. Nature Methods, 12 (2015) 959-962. Link.
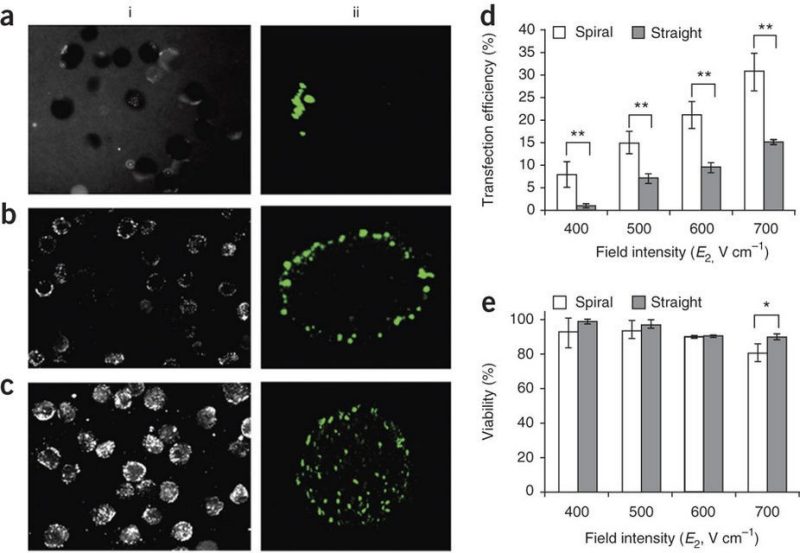
Transfection of cells using flow-through electroporation based on constant voltage
Nature Protocols 2011
Geng, T., Zhan, Y., Wang, J., Lu, C. Nature Protocols 6 (2011) 1192-1208. Link.
Publications
(# denotes equal contribution, * denotes corresponding authors)
2026
- Hadlock, T. M., Neice, J. D., Li, S., Mor, S., Lu, C.*
A microreactor system for point-of-care viral genome sequencing. Biosensors and Bioelectronics 297 (2026) 118369. Link
2025
- Wu, P.#, Liu, Z.#, Zheng, L.#, Du, Y.#, Zhou, Z., Wang, W., Lu, C.*
Comprehensive multimodal and multiomic profiling reveals epigenetic and transcriptional reprogramming in lung tumors. Communications Biology 8 (2025) 527. Link
- Jaster, A. M., Hadlock, T. M.#, Buzzi, B.#, Maltman, J. L., Silva, G. M., Saha, S., Koseli, E., Pondelick, A. M., Thakur, N., Zhang, X., Li, G., Ledesma-Corvi, S., Moore, K. N., Peterson, H. R., Fujita, B., Zylko, A. L., Lewis, M. R., Poklis, J. L., Halquist, M. S., Wolstenholme, J. T., Selley, D. E., Hamilton, P. J., Lu, C.*, Damaj, M. I.*, González-Maeso, J.*
Sex-specific role of the 5-HT2A receptor in psilocybin-induced extinction of opioid reward. Nature Communications 16 (2025) 10206. Link
2024
- Udayasuryan, B.#, Zhou, Z.#, Ahmad, R.N., Sobol, P., Deng, C., Nguyen, T. T. D., Kodikalla, S., Morrison, R., Goswami, I., Slade, D. J., Verbridge, S., Lu, C.*
Fusobacterium nucleatum infection modulates the transcriptome and epigenome of HCT116 colorectal cancer cells in an oxygen-dependent manner. Communications Biology 7 (2024) 551.Link
Drug Discovery News
“No oxygen? No problem for these cancer-aiding bacteria”
- Liu, Z.#, Deng, C.#, Zhou, Z., Xiao, Y., Jiang, S., Zhu, B., Naler, L., Jia, X., Yao, D., Lu, C.*
Epigenomic tomography for probing spatially defined chromatin state in the brain. Cell Reports Methods 4 (2024) 100738.Link
VT News
“New spatial profiling approach maps out discoveries for future brain research”
The Scientist
“An Epigenetic Brain Scan”
2023
- Zhu, B.#, Ainsworth, R.J.#, Wang, Z., Liu, Z., Sierra, S., Deng, C., Callado, L.F., Meana, J.J., Wang, W.*, Lu, C.*, Gonzalez-Maeso, J*.
Antipsychotic-induced epigenomic reorganization in frontal cortex of individuals with schizophrenia. eLife 12 (2023) RP92393. Link
- Zhang, Q., Ma, S., Liu, Z., Zhu, B., Zhou, Z., Li, G., Meana, J., González-Maeso, J., Lu, C.*
Droplet-based bisulfite sequencing for high-throughput profiling of single-cell DNA methylomes. Nature Communications 14 (2023) 4672. Link
2022
- Afrose, S., Song, W., Nemeroff, C.B., Lu, C.*, Yao, D.*
Subpopulation-specific Machine Learning Prognosis for Underrepresented Patients with Double Prioritized Bias Correction. Communications Medicine 2 (2022) 111. Link
VT News
“New AI fairness technique has significant lifesaving implications”
- Liu, Z., Naler, L.B., Zhu, Y., Deng, C., Zhang, Q., Zhu, B., Zhou, Z., Sarma, M., Murray, A., Xie, H., Lu, C.*
nMOWChIP-seq: low-input genome-wide mapping of non-histone targets. NAR Genomics and Bioinformatics 4 (2022) Iqac030. Link
- Hsieh, Y.-P. #, Naler, L.B. #, Ma, S., , Lu, C.*
Cell-type-specific epigenomic variations associated with BRCA1 mutation in pre-cancer human breast tissues. NAR Genomics and Bioinformatics 4 (2022) Iqac006. Link
- Naler, L.B. #, Hsieh, Y.-P. #, Geng, Z., Zhou, Z., Li, L.*, Lu, C.*
Epigenomic and transcriptomic analyses reveal differences between low-grade inflammation and severe exhaustion in LPS-challenged murine monocytes. Communications Biology 5 (2022) 102. Link2021
- de la Fuente Revenga, M., Zhu, B. #, Guevara, C.A. #, Naler, L.B., Saunders, J.M., Zhou, Z., Toneatti, R., Sierra, S., Wolstenholme, J.T., Beardsley, P.M., Huntley, G.W., Lu, C.*, Gonzalez-Maeso, J.*
Prolonged epigenomic and synaptic plasticity alterations following single exposure to a psychedelic in mice. Cell Reports 37 (2021) 109836. Link
VCU School of Medicine News
“Single dose of a psychedelic drug alters neurons’ structure and gene environment”
Outlook at Nature
“Epigenetic roots of long-lasting therapy”
BioTechniques
“Are psychedelics superior for treating mental illness? Potentially, yes”
VT News
“Psychedelics show promise in treating mental illness”
Selected for cover:
2020
- Deng, C., Murphy, T., Zhang, Q., Naler, L., Xu, A., Lu, C.*
Multiplexed and Ultralow-input ChIP-seq enabled by tagmentation-based indexing and facile microfluidics. Analytical Chemistry 92 (2020) 13661 - 13666. Link
- Murphy, T. W., Hsieh, Y.-P., Zhu, B., Naler, L.B., Lu, C.*
Microfluidic platform for next-generation sequencing library preparation with low-input samples. Analytical Chemistry 92 (2020) 2519 - 2526. Link
- Rahtes, A., Pradhan, K., Sarma, M., Xie, H., Lu, C.* Li, L.*
Phenylbutyrate facilitates homeostasis of non-resolving inflammatory macrophages. Innate Immunity 26 (2020) 62-72. Link2019
- Zhu, B. #, Hsieh, Y.-P. #, Murphy, T. W., Zhang, Q., Naler, L. B., Lu, C.*
MOWChIP-seq for low-input and multiplexed profiling of genome-wide histone modifications. Nature Protocols 14 (2019) 3366-3394. Link
Included in the National Cancer Institute's Research Highlights for the Epidemiology and Genomics Research Program for 2019
VT News
"Highly sensitive epigenomic technology combats disease."
- Deng, C. #, Naler, L. B. #, Lu, C.*
Microfluidic epigenomic mapping technologies for precision medicine. Lab on a Chip 19 (2019) 2630-2650. Link
- Hu, Y., Xu, F., Zhang, R., Legarda, D., Dai, J., Wang, D., Li, H., Zhang, Y., Xue, Q., Dong, G., Zhang, H., Lu, C., Mortha, A., Liu, J., Cravedi, P., Ting, A., Li, L., Qi, C.-f., Pierce, S., Merad, M., Heeger, P., Xiong, H.
Interleukin-1β-induced IRAK1 ubiquitination is required for TH-GM-CSF cell differentiation in T cell-mediated inflammation. Journal of Autoimmunity 102 (2019) 50-64. Link
- Zhang, X., Wang, Y., Chiang, H.-C., Hsieh, Y.-P., Lu, C., Park, B. H., Jatoi, I., Jin, V. X., Hu, Y., Li, R.
BRCA1 mutations attenuated super-enhancer function and chromatin looping in haploinsufficient human breast epithelial cells. Breast Cancer Research 21 (2019) 51. Link
- Sarma, M., Lee, J., Ma, S., Li, S., Lu, C.*
A diffusion-based microfluidic device for single-cell RNA-seq. Lab on a Chip 19 (2019) 1247-1256. Link
- Cox, M. #, Deng, C. #, Naler, L., Lu, C.*, Verbridge, S.*
Effects of culture condition on epigenomic profiles of brain tumor cells. ACS Biomaterials Science & Engineering 5 (2019) 1554-1552. Link
- Murphy, T.W., Sheng, J., Naler, L.B., Feng, X., Lu, C.*
On-chip Manufacturing of Synthetic Proteins for Point-of-care Therapeutics. Microsystems & Nanoengineering 5 (2019) 13. Link
Phys.org News Feature
“Therapeutics-on-a-chip (TOC): Manufacturing synthetic proteins for point-of-care therapeutics”
- Zhu, Y., Cao, Z., Lu, C.*
Microfluidic MeDIP-seq for low-input methylomic analysis of mammary tumorigenesis in mice. Analyst 144 (2019) 1904-1915. Link
Selected for cover:
2018
- Murphy, T.W., Hsieh, Y.-P., Ma, S., Zhu, Y., Lu, C.*
Microfluidic low-input fluidized-bed enabled ChIP-seq device for automated and parallel analysis of histone modifications. Analytical Chemistry 90 (2018) 7666–7674. Link
- Ma, S., Hsieh, Y.-P., Ma, J., Lu, C.*
Low-input and multiplexed microfluidic assay reveals epigenomic variation across cerebellum and prefrontal cortex. Science Advances 4 (2018) eaar8187. Link
- Ma, S., de la Fuente Revenga, M., Sun, Z., Sun, C., Murphy, T.W., Xie, H., Gonzalez-Maeso, J., Lu, C.*
Cell-type-specific brain methylomes profiled via ultralow-input microfluidics. Nature Biomedical Engineering 2 (2018) 183-194. Link; free view-only copy.
News & Views by Feng-Mao Lin, Shu Chien and Zhen Chen.
“Sequencing technology: Epigenetic profiling with ultralow DNA amounts"; free view-only copy ;
Behind-the-paper post by Chang Lu.
“Using microfluidics to map brain epigenomes”
VT news
"Epigenomic tool breakthrough has implications for identifying disease processes".
- Sun, C., Lu, C.*
Microfluidics-based chromosome conformation capture (3C) technology for examining chromatin organization with a low quantity of cells. Analytical Chemistry 90 (2018) 3714–3719. Link
- Murphy, T.W., Zhang, Q., Naler, L.B., Ma, S., Lu, C.*
Recent advances on microfluidic technologies for single cell analysis. (review) Analyst 143 (2018) 60-80. Link
Included in Analyst 2018 "Most Downloaded Articles" Themed Collection
2017
- Ma, S., Murphy, T.W., Lu, C.*
Microfluidics for genome-wide studies involving next generation sequencing. (review) Biomicrofluidics 11 (2017) 021501. Link
- Sun, C., Hsieh, Y.P., Ma, S., Geng, S., Cao, Z., Li, L., Lu, C.*
Immunomagnetic Separation of Tumor Initiating Cells by Screening Two Surface Markers. Scientific Reports 7 (2017) 40632. Link
2016
- Lu, C., Verbridge, S.S.(ed.)
Microfluidic Methods for Molecular Biology. Springer, 2016. ISBN 978-3-319-30017-7.
- Zhu, Y., Lu, C.*
Microfluidic Chromatin Immunoprecipitation for Analysis of Epigenomic Regulations. Microfluidic Methods for Molecular Biology. (Lu, C., Verbridge, S.S. ed.) Springer, 2016.
- Sun, C., Hassanisaber, H., Yu, R., Ma, S., Verbridge, S.S., Lu, C.*
Paramagnetic structures within a microfluidic channel for enhanced immunomagnetic isolation and surface patterning of cells. Scientific Reports 6 (2016) 29407. Link
- Ma, S., Bryson, B.D., Sun, C., Fortune, S.M., Lu, C.*
RNA extraction from a mycobacterium under ultrahigh electric field intensity in a microfluidic device. Analytical Chemistry 88 (2016) 5053-5057. Link
- Cao, Z., Lu, C.*
A microfluidic device with integrated sonication and immunoprecipitation for sensitive epigenetic assays. Analytical Chemistry 88 (2016) 1965-1972. Link
2015
- Cao, Z., Chen, C., He, B., Tan, K., Lu, C.*
A microfluidic device for epigenomic profiling using 100 cells. Nature Methods, 12 (2015) 959-962. Link
- Cao, Z., Lu, C.*
Quantitative detection of nucleocytoplasmic transport of native proteins at the single cell level. Single cell protein analysis, Methods in Molecular Biology, (Singh, A.K. and Chandrasekaran, A. ed.) Springer, 2015.
2014
- del Rosal, B., Sun, C., Yan, Y., Mackenzie, M.D., Lu, C., Bettiol, A.A., Kar, A.K., Jaque, D.
Flow effects in the laser-induced thermal loading of optical traps and optofluidic devices. Optics Express 22 (2014) 23938-23954. Link
- Sun, C., Ouyang, M., Cao, Z., Ma, S., Alqublan, H., Sriranganathan, N., Wang, Y., Lu, C.*
Electroporation-delivered fluorescent protein biosensors for probing molecular activities in cells without genetic encoding. Chemical Communications 50 (2014) 11536-11539. Link
- Sun, C., Cao, Z., Wu, M., Lu, C.*
Intracellular Tracking of Single Native Molecules with Electroporation-delivered Quantum Dots. Analytical Chemistry 86 (2014) 11403-11409. Link
- Ma, S., Loufakis, D.N., Cao, Z., Chang, Y., Achenie, L.E.K., Lu, C.*
Diffusion-based Microfluidic PCR for “One-pot” Analysis of Cells. Lab on a Chip 14 (2014) 2905-2909. Link
Selected as HOT article and the journal cover (issue 16):
- Ma, S., Schroeder, B., Sun, C., Loufakis, D.N., Cao, Z., Sriranganathan, N., Lu, C.*
Electroporation-based delivery of cell-penetrating peptide conjugates of peptide nucleic acids for antisense inhibition of intracellular bacteria. Integrative Biology 6 (2014) 973-978. Link
- Loufakis, D.N., Cao, Z., Ma, S., Mittelman, D., Lu, C.*
Focusing of mammalian cells under an ultrahigh pH gradient created by unidirectional electropulsation in a confined microchamber. Chemical Science 5 (2014) 3331-3337. Link
- Cao, Z., Geng, S., Li, L., Lu, C.*
Detecting intracellular translocation of native proteins quantitatively at the single cell level. Chemical Science 5 (2014) 2530-2535. Link
VT news (May 13, 2014): Researchers follow a protein's travel inside cells to improve patient monitoring, develop drugs.
2013
- Geng, T., Lu, C.*
Microfluidic Electroporation for Cellular Analysis and Delivery. (Critical review) Lab on a Chip 13 (2013) 3803-3821. Link
- del Rosal, B., Sun, C., Loufakis, D.N., Lu, C., Jaque, D.
Thermal loading in flow-through electroporation microfluidic devices. Lab on a Chip 13 (2013) 3119-3127. Link
- Cao, Z., Chen, F., Bao, N., He, H., Xu, P., Jana, S., Jung, S., Lian, H., Lu, C.*
Droplet Sorting Based on the Number of Encapsulated Particles Using a Solenoid Valve. Lab on a Chip 13 (2013) 171-178. Link
2012
- Geng, T., Bao, N., Sriranganathan, N., Li, L., Lu, C.*
Genomic DNA extraction from cells by electroporation on an integrated microfluidic platform. Analytical Chemistry 84 (2012) 9632-9639. Link
- Zhan, Y., Sun, C., Cao, Z., Bao, N., Xing, J., Lu, C.*
Release of intracellular proteins by electroporation with preserved cell viability. Analytical Chemistry 84 (2012) 8102-8105. Link
- Zhan, Y., Loufakis, D.N., Bao, N., Lu, C.*
Characterizing osmotic lysis kinetics under microfluidic hydrodynamic focusing for erythrocyte fragility studies. Lab on a Chip 12 (2012) 5063-5068. Link
- Zhan, Y., Lu, C.
Microfluidic devices for cellular proteomics studies. Microfluidic technologies for human health. (Demirci, U. ed.), Chapter 8. World Scientific Publishing Co, Singapore, 2012.
- Shi, C., Shan, X., Pan, Z.-Q., Xu, J.J., Lu, C., Bao, N., Gu, H.-Y.
Quantum-Dot-Modified Carbon Tape Electrodes for Reproducible Electrochemiluminescence Emission on a Paper-based Platform. Analytical Chemistry 84 (2012) 3033-3038. Link
- Zhan, Y., Cao, Z., Bao, N., Li, J., Wang, J., Geng, T., Lin, H., Lu, C.*
Low-frequency ac electroporation shows strong frequency dependence and yields comparable transfection results to dc electroporation. Journal of Controlled Release 160 (2012) 570-576. Link
- Awwad, Y., Geng, T., Baldwin, A.S., Lu, C.*
Observing single cell NF-kappa B dynamics under stimulant concentration gradient. Analytical Chemistry 84 (2012) 1224-1228. Link
2011
 Wang, J., Zhan, Y., Bao, N., Lu, C.*
Wang, J., Zhan, Y., Bao, N., Lu, C.*
Quantitative measurement of quantum dot uptake at the cell population level using microfluidic evanescent-wave-based flow cytometry. Lab on a Chip 12 (2012) 1441-1445. Link
Selected as the journal cover (issue 8):
- Chen, F., Zhan, Y., Geng, T., Lian, H., Xu, P., Lu, C.*
Chemical transfection of cells in picoliter aqueous droplets in fluorocarbon oil. Analytical Chemistry 83 (2011) 8816-8820. Link
- Bao, N., Kodippili, G.C., Giger, K.M., Fowler, V.M., Low, P.S., Lu, C.*
Single-cell Electrical Lysis of Erythrocytes Detects Deficiencies in the Cytoskeletal Protein Network. Lab on a Chip 11 (2011) 3053-3056. Link
Selected as a HOT article.
- Geng, T., Bao, N., Litt, M.D., Glaros, T.G., Li, L., Lu, C.*
Histone modification analysis by chromatin immunoprecipitation from a low number of cells on a microfluidic platform. Lab on a Chip 11 (2011) 2842-2848. Link
- Geng, T., Zhan, Y., Wang, J., Lu, C.*
Transfection of cells using flow-through electroporation based on constant voltage. Nature Protocols 6 (2011) 1192-1208. Link
2010
- Wang, J., Fei, B., Zhan, Y., Geahlen, R.L. and Lu, C.*
Kinetics of NF-kappaB nucleocytoplasmic transport probed by single-cell screening without imaging. Lab on a Chip 10 (2010) 2911 - 2916. Link
- Wang, J., Fei, B., Geahlen, R.L. and Lu, C.*
Quantitative analysis of protein translocations by microfluidic total internal reflection fluorescence flow cytometry. Lab on a Chip 10 (2010) 2673-2679. Link
- Zhan, Y., Martin, V.A., Geahlen, R.L. and Lu, C.*
One-step extraction of subcellular proteins from eukaryotic cells. Lab on a Chip 10 (2010) 2046-2048. Link
- Geng, T., Zhan, Y., Wang, H.Y., Witting, S.R., Cornetta, K.G. and Lu, C.*
Flow-through electroporation based on constant voltage for large-volume transfection of cells. Journal of Controlled Release 144 (2010) 91-100. Link
- Wang, J., Zhan, Y., Ugaz, V.M. and Lu, C.*
Vortex-assisted DNA Delivery. Lab on a Chip 10 (2010) 2057-2061. Link
Research highlight in Nature (Vol 466, July 8 2010): "Biotechnology: Swirling cells."
Update in Chemical Engineering Progress (CEP) (August 2010): A New Spin on DNA Delivery.
Virginia Tech News (July 12, 2010): A new spin on gene delivery.
Top 10 most accessed articles of Lab Chip in July and August 2010.
Selected as the journal cover (issue 16):
- Bao, N., Le, T.T., Cheng, J.X. and Lu, C.*
Microfluidic electroporation of tumor and blood cells: observation of nucleus expansion and implications on selective analysis and purging of circulating tumor cells. Integrative Biology 2 (2010) 113-120. Link
Highlighted in Highlights in Chemical Biology (Jan 13, 2010): Tackling rogue tumour cells.
Top 10 articles (2010). Selected as the journal cover:
- Lu, C. (ed.)
Chemical Cytometry: Ultrasensitive analysis of single cells. Wiley-VCH, Weinheim, Germany, Jan 2010. ISBN: 978-3-527-32495-8.
- Lu, C., Wang, J., Bao, N. and Wang, H.Y.
Electroporative flow cytometry for single cell analysis. Chemical Cytometry: Ultrasensitive analysis of single cells. (Lu, C. ed.) Wiley-VCH, Weinheim, Germany, 2010.
2009
- Zhan, Y., Wang, J., Bao, N. and Lu, C.*
Electroporation of cells in microfluidic droplets. Analytical Chemistry 81 (2009) 2027-2031. Link
Selected for inclusion in "Microfluidics thematic collection" in Analytical Chemistry (Oct 2010).
- Geng, T., Bao, N., Gall, O.Z. and Lu, C.*
Modulating DNA adsorption on silica beads using an electrical switch. Chemical Communications (2009) 800-802. Link
2008
- Wang, J., Bao, N., Paris, L.L., Geahlen, R.L. and Lu, C.
Total internal reflection fluorescence flow cytometry. Analytical Chemistry 80 (2008) 9840-9844. Link
- Bao, N., Zhan, Y. and Lu, C.
Microfluidic electroporative flow cytometry for studying single cell biomechanics. Analytical Chemistry 80 (2008) 7714-7719. Link
Highlighted by Purdue News (Sept 18, 2008), Journal and Courier (Oct 5, 2008), and The Exponent (Oct 15, 2008).
- Wang, H.Y., Bao, N. and Lu, C.
A microfluidic cell array with individually addressable culture chambers. Biosensors & Bioelectronics 24 (2008) 613-617. Link
- Wang, H.W., Bao, N., Le, T.T., Lu, C. and Cheng, J.X.
Microfluidic CARS cytometry. Optics Express 16 (2008) 5782-5789. Link
Highlighted in Biophotonics International (Vol 15, Issue 6, 2008): CARS flow cytometry on a chip.
- Bao, N., Wang, J. and Lu, C.
Recent advances in electric analysis of cells in microfluidic systems. (Invited review) Analytical and Bioanalytical Chemistry 391 (2008) 933-942. Link
- Bao, N. and Lu, C.
A microfluidic device for physical trapping and electrical lysis of bacterial cells. Applied Physics Letters 92 (2008) 214103. Link
Selected for inclusion in Virtual Journal of Nanoscale Science & Technology and Virtual Journal of Biological Physics Research.
- Bao, N., Wang, J. and Lu, C.
Microfluidic electroporation for selective release of intracellular molecules at the single cell level. Electrophoresis 29 (2008) 2939-2944. Link
- Bao, N., Jagadeesan, B., Bhunia, A.K., Yao, Y., and Lu, C.
Quantification of bacterial cells based on autofluorescence on a microfluidic platform. Journal of Chromatography A 1181 (2008) 153-158. Link
- Wang, H.Y. and Lu, C.
Microfluidic electroporation for delivery of small molecules and genes into cells using a common dc power supply. Biotechnology and Bioengineering 100 (2008) 579-586. Link
- Wang, F., Wang, H., Wang, J., Wang, H.Y., Rummel, P.L., Garimella, S.V. and Lu, C.
Microfluidic delivery of small molecules into mammalian cells based on hydrodynamic focusing. Biotechnology and Bioengineering 100 (2008) 150-158. Link
Selected as the journal cover:
- Wang, J., Bao, N., Paris, L.L., Wang, H.Y., Geahlen, R.L. and Lu, C.
Detection of kinase translocation using microfluidic electroporative flow cytometry. Analytical Chemistry 80 (2008) 1087-1093. Link
Highlighted by Purdue News (Jan 22, 2008) "New technique quickly detects cancer indicator"; Proteomonitor (Vol 8, Number 5, Jan 31, 2008) "Purdue team develops method for detecting kinase translocation"; Biophotonics International (Vol 15, Number 3, 2008) "Flow cytometry technique detects translocating proteins"; and Biocompare Technology Spotlight (Jul 14, 2008) "Single cell analysis".
- Bao, N. and Lu, C.
Microfluidics-based lysis of bacteria and spores for detection and analysis. Principles of Bacterial Detection: Biosensors, Recognition Receptors and Microsystems (Zourob, M. et al. ed.), P783-796, Springer, New York, NY, 2008.
2007
- Wang, J., Stine, M.J. and Lu, C.
Microfluidic cell electroporation using a mechanical valve. Analytical Chemistry 79 (2007) 9584-9587. Link
- Wang, J. and Lu, C.
Single molecule lamda-DNA stretching studied by microfluidics and single particle tracking. Journal of Applied Physics 102 (2007) 074703. Link
- Wang, H.Y., Banada, P.P., Bhunia, A.K. and Lu, C.
Rapid electrical lysis of bacterial cells in a microfluidic device. Methods in Molecular Biology, Vol. 385: Microchip-based Assay Systems: Methods and Applications (P.N. Floriano ed.), P23-35, Humana Press, Totowa, NJ, 2007.
2006
- Wang, J. and Lu, C.
Microfluidic cell fusion under continuous direct current voltage. Applied Physics Letters 89 (2006) 234102. Link
Highlighted in Purdue News (Dec 4, 2006) "The power of one: A simpler, cheaper method for cell fusion."
Selected for inclusion in Virtual Journal of Biological Physics Research.
- Wang, H.Y. and Lu, C.
Microfluidic chemical cytometry based on modulation of local field strength. Chemical Communications (2006) 3528-3530. Link
Selected for inclusion in RSC Chemical Biology Virtual Journal.
- Wang, H.Y. and Lu, C.
High-throughput and real-time study of single cell electroporation using microfluidics: effects of medium osmolarity. Biotechnology & Bioengineering 95 (2006) 1116-1125. Link
- Wang, H.Y. and Lu, C.
Electroporation of mammalian cells in a microfluidic channel with geometric variation. Analytical Chemistry 78 (2006) 5158-5164. Link
Highlighted in Purdue News (June 21, 2006) "Microchannels, electricity aid drug discovery, early diagnosis"; Bioscience Technology (Vol 31, Number 8, Aug 2006) "The Other Chips: Microfluidic Devices Come of Age"; and Medical Device & Diagnostic Industry (Vol 28, Number 9, Sept 2006)"Single-cell Analysis: Quick and Easy Detection"
- Wang, H.Y., Bhunia, A.K. and Lu, C.
A microfluidic flow-through device for high throughput electrical lysis of bacterial cells based on continuous DC voltage. Biosensors & Bioelectronics 22 (2006) 582-588. Link
- Chu, K.L., Gold, S., Subramanian, R., Lu, C., Shannon, M.A. and Masel, R.I.
A nanoporous silicon membrane electrode assembly for on-chip micro fuel cell applications. Journal of Microelectromechanical Systems 15 (2006) 671-677. Link
2005 and before
- Lu, C., Smith, A.E. and Craighead, H.G.
Separation of denatured proteins in free solution on a microchip based on differential binding of alkyl sulfates with different carbon chain lengths. Chemical Communications (2005) 183-185. Link
Selected for inclusion in RSC Chemical Biology Virtual Journal.
- Gold, S., Chu, K.L., Lu, C., Shannon, M.A. and Masel, R.I.
Acid loaded porous silicon as a proton exchange membrane for micro fuel cells. Journal of Power Sources 135 (2004) 198-203. Link
- Waszczuk, P., Lu, G.Q., Wieckowski, A., Lu, C., Rice, C. and Masel, R.I.
UHV and electrochemical studies of CO and methanol adsorbed at platinum/ruthenium surfaces, and reference to fuel cell catalysis. Electrochimica Acta 47 (2002) 3637-3652 (Invited review). Link
- Lu, C., Rice, C., Masel, R.I., Babu, P.K., Waszczuk, P., Kim, H.S., Oldfield, E. and Wieckowski, A.
UHV, electrochemical NMR, and electrochemical studies of platinum/ruthenium fuel cell catalysts. Journal of Physical Chemistry B 106 (2002) 9581-9589. Link
- Lu, C., Lee, I.C., Masel, R.I., Wieckowski, A. and Rice, C.
Correlations between the heat of adsorption and the position of the center of the d-band: differences between computation and experiment. Journal of Physical Chemistry A 106 (2002) 3084-3091. Link
- Lu, C. and Masel, R.I.
The effect of ruthenium on the binding of CO, H2 and H2O on Pt(110). Journal of Physical Chemistry B 105 (2001) 9793-9797. Link
- Lu, C., Thomas, F.S. and Masel, R.I.
Chemistry of methoxonium on (2x1)Pt(110). Journal of Physical Chemistry B 105 (2001) 8583-8590. Link
- Thomas, F.S., Lu, C., Lee, I.C., Chen, N.S. and Masel, R.I.
Evidence for a cation intermediate during methanol dehydration on Pt(110). Catalysis Letters 72 (2001) 167-175. Link
- Ali, N., Lu, C. and Masel, R.I.
Catalytic oxidation of odorous organic acids. Catalysis Today 62 (2000) 347-353. Link

