Ultralow-input epigenomic analysis
We have developed microfluidic devices with ultrahigh sensitivity and simple structures for epigenetic/epigenomic analysis of cells. We demonstrated microfluidic ChIP-qPCR based on 50 cells and MOWChIP-seq using 100 cells. MOWChIP-seq was the first microfluidic technologies for mapping epigenomes (2015 Nat Methods). We also developed MID-RRBS for profiling DNA methylomes with nanogram-to-single-cell quantities of DNA (2018 Nat Biomed Eng) and SurfaceChIP-seq for profiling histone modifications with both ultralow-input (30 cells per assay) and high throughput (8 assays in parallel) (2018 Sci Adv). These assays paved the way for testing scarce primary tissues from lab animals and patients with direct biomedical relevance.
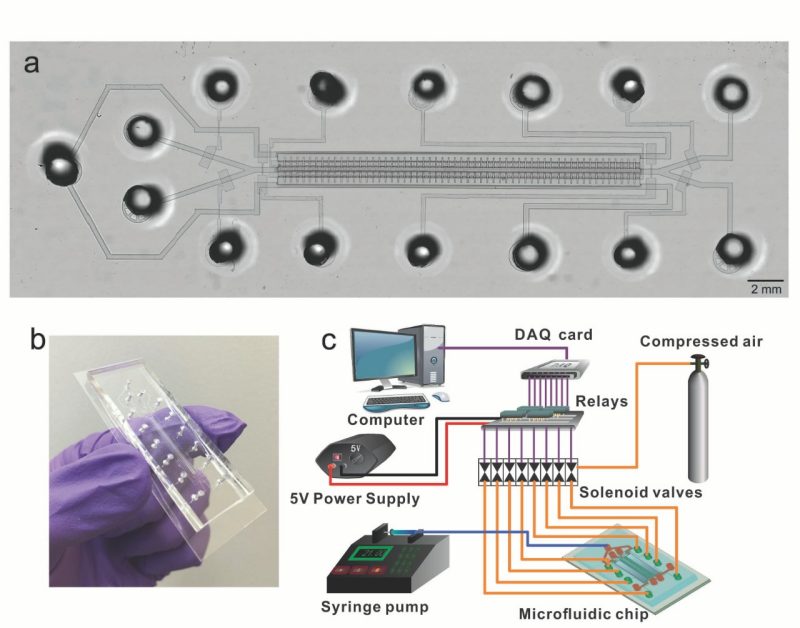
Figure 1. Design and setup of a single-unit MID-RRBS device. (Ma, S. et al. Nat BME 2018).
Electroporation for gene delivery
We developed a unique flow-through electroporation technology working under constant voltage for delivering genes via the entire membrane surface and handling a large volume of cell sample. This technology was highlighted by Nature and widely adapted for gene delivery applications by researchers around the world.
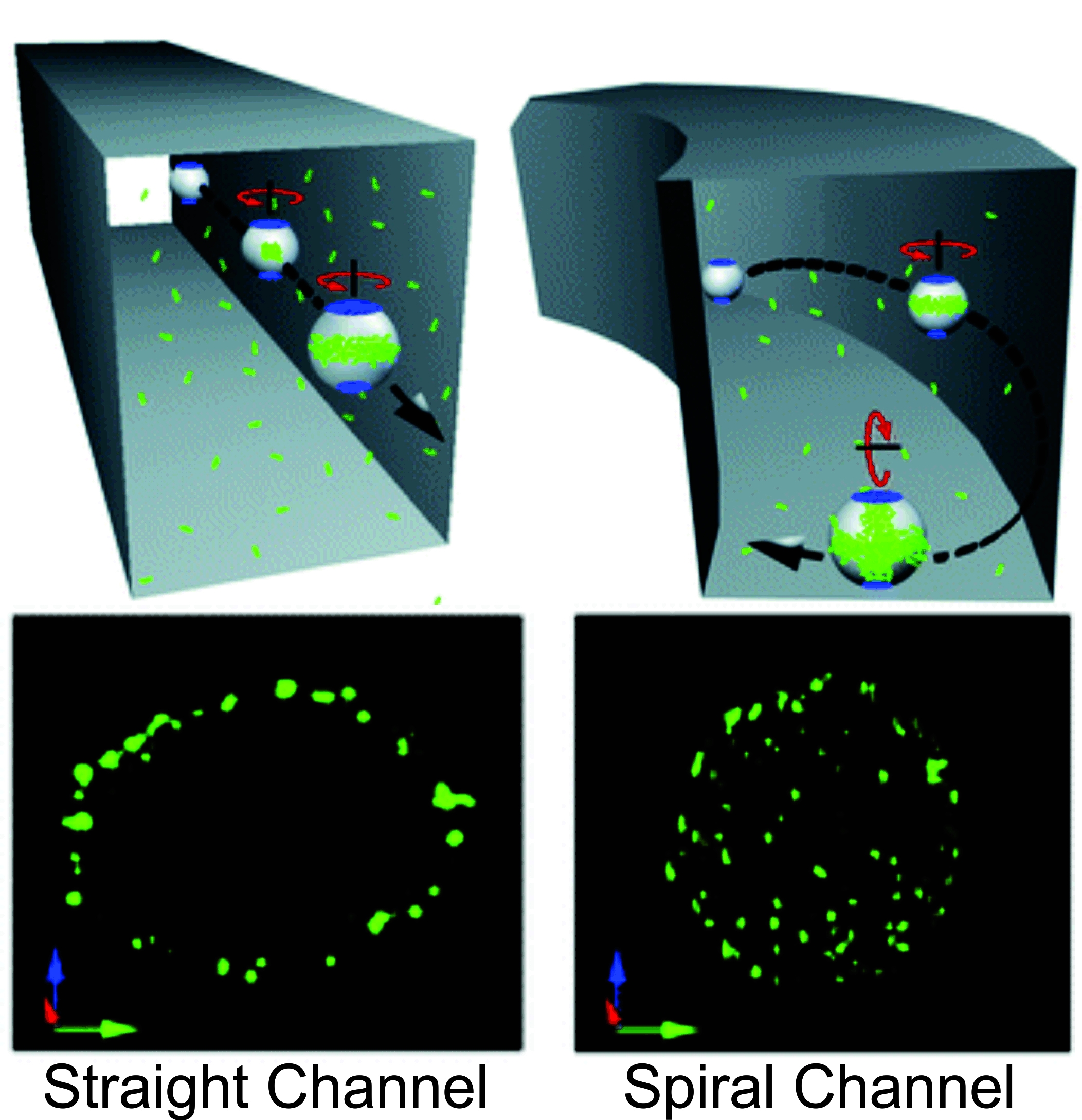
Figure 2. Demonstration of vortex assisted DNA delivery in a straight channel and curved path harnessing Dean flows. (Wang, J. et al. Lab Chip 2010).
Single cell analysis
When heterogeneous cell populations, such as those derived from patients, are studied, most sample preparation methods call for both genetic and protein contents to be averaged over the tissue samples. The data yielded from these experiments often do not reflect the properties of cell subsets or provide information on kinetics of biological processes. Microfluidic devices offer new opportunities for manipulation and analysis at the single cell level. We develop novel microfluidic cytometry tools for obtaining unique information on cells such as the subcellular location of an intracellular protein, cell deformability, and cell surface events with high throughput and single cell resolution. These tools include microfluidic chemical cytometry, electroporative flow cytometry, and total internal reflection fluorescence flow cytometry. We envision that these techniques will find applications to mechanistic studies of molecular/cell biology, drug discovery, and clinical diagnosis/prognosis.
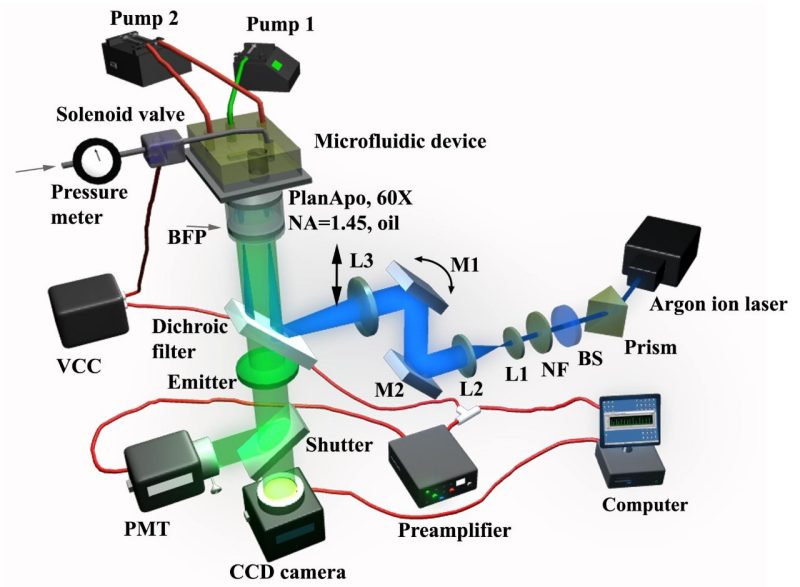
Figure 3. The setup of the total internal reflection fluorescence flow cytometer (TIRF-FC) (Wang, J. et al. Anal. Chem. 2008).
Droplet microfluidics
We developed various droplet microfluidic devices for cell transfection, analysis and sorting. We developed the first droplet sorter that enriched a subset of droplets containing the same number of particles/cells.
Figure 4. The sorting of two-cell droplets from a mixed droplet population. The video was recorded at a frame rate of 15 fps and is played at 5 fps. The flow rates used for the aqueous phase and the oil phase were 0.15 and 3.7 µl/min, respectively. (Cao, Z. et al. Lab Chip 2013).

